Getting Started¶
Note
A cookbook of simple recipes for advanced data analytics using PySD is available at: http://pysd-cookbook.readthedocs.org/
The cookbook includes models, sample data, and code in the form of iPython notebooks that demonstrate a variety of data integration and analysis tasks. These models can be executed on your local machine, and modified to suit your particular analysis requirements.
Importing a model and getting started¶
To begin, we must first load the PySD module, and use it to import a model file:
>>> import pysd
>>> model = pysd.read_vensim('Teacup.mdl')
This code creates an instance of the PySD Model class from an example model that we will use as the system dynamics equivalent of ‘Hello World’: a cup of tea cooling at room temperature.
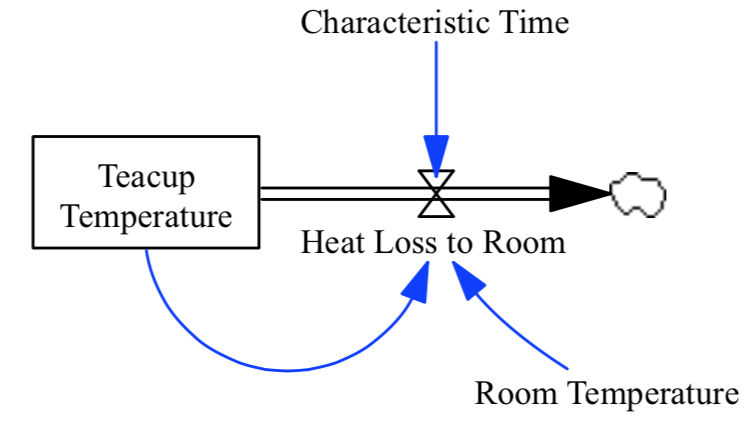
Note
The teacup model can be found in the samples of the test-models repository.
To view a synopsis of the model equations and documentation, use the doc property of the Model class. This will generate a listing of all model elements, their documentation, units, and initial values, where appropriate, and return them as a pandas.DataFrame. Here is a sample from the teacup model:
>>> model.doc
Real Name Py Name Subscripts Units Limits Type Subtype Comment
0 Characteristic Time characteristic_time None Minutes (0.0, nan) Constant Normal How long will it take the teacup to cool 1/e o...
1 FINAL TIME final_time None Minute (nan, nan) Constant Normal The final time for the simulation.
2 Heat Loss to Room heat_loss_to_room None Degrees Fahrenheit/Minute (nan, nan) Auxiliary Normal This is the rate at which heat flows from the ...
3 INITIAL TIME initial_time None Minute (nan, nan) Constant Normal The initial time for the simulation.
4 Room Temperature room_temperature None Degrees Fahrenheit (-459.67, nan) Constant Normal Put in a check to ensure the room temperature ...
5 SAVEPER saveper None Minute (0.0, nan) Auxiliary Normal The frequency with which output is stored.
6 TIME STEP time_step None Minute (0.0, nan) Constant Normal The time step for the simulation.
7 Teacup Temperature teacup_temperature None Degrees Fahrenheit (32.0, 212.0) Stateful Integ The model is only valid for the liquid phase o...
8 Time time None None (nan, nan) None None Current time of the model.
Note
You can also load an already translated model file. This will be faster than loading an original model, as the translation is not required:
>>> import pysd
>>> model = pysd.load('Teacup.py')
Note
The functions pysd.read_vensim(), pysd.read_xmile() and pysd.load() have optional arguments for advanced usage. You can check the full description in Model loading or using help() e.g.:
>>> import pysd
>>> help(pysd.load)
Note
Not all the features and functions are implemented. If you are in trouble while importing a Vensim or Xmile model check the Vensim supported functions or Xmile supported functions.
Running the Model¶
The simplest way to simulate the model is to use the run() command with no options. This runs the model with the default parameters supplied in the model file, and returns a pandas.DataFrame of the values of the model components at every timestamp:
>>> stocks = model.run()
>>> stocks
Characteristic Time Heat Loss to Room Room Temperature Teacup Temperature FINAL TIME INITIAL TIME SAVEPER TIME STEP
0.000 10 11.000000 70 180.000000 30 0 0.125 0.125
0.125 10 10.862500 70 178.625000 30 0 0.125 0.125
0.250 10 10.726719 70 177.267188 30 0 0.125 0.125
0.375 10 10.592635 70 175.926348 30 0 0.125 0.125
0.500 10 10.460227 70 174.602268 30 0 0.125 0.125
... ... ... ... ... ... ... ... ...
29.500 10 0.565131 70 75.651312 30 0 0.125 0.125
29.625 10 0.558067 70 75.580671 30 0 0.125 0.125
29.750 10 0.551091 70 75.510912 30 0 0.125 0.125
29.875 10 0.544203 70 75.442026 30 0 0.125 0.125
30.000 10 0.537400 70 75.374001 30 0 0.125 0.125
[241 rows x 8 columns]
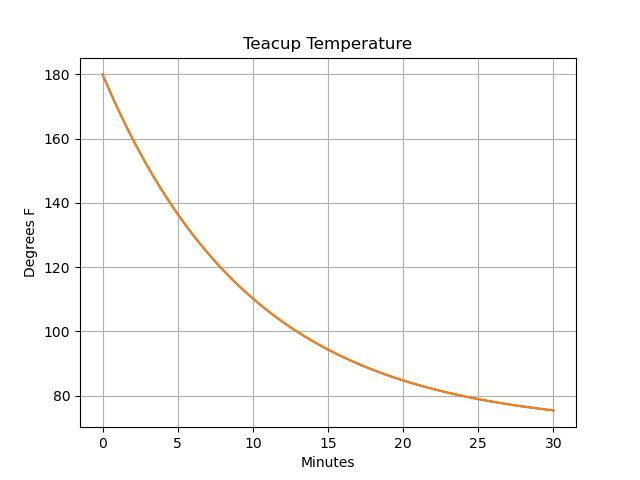
Pandas proovides a simple plotting capability, that we can use to see how the temperature of the teacup evolves over time:
>>> import matplotlib.pyplot as plt
>>> stocks["Teacup Temperature"].plot()
>>> plt.title("Teacup Temperature")
>>> plt.ylabel("Degrees F")
>>> plt.xlabel("Minutes")
>>> plt.grid()
To show a progressbar during the model integration, the progress argument can be passed to the run() method:
>>> stocks = model.run(progress=True)
Note
The full description of the run() method and other methods can be found in the Model methods section.
Running models with DATA type components¶
Venim allows to import DATA type data from binary .vdf files. Variables defined without an equation in the model, will attempt to read their values from the .vdf. PySD allows running models with this kind of data definition using the data_files argument when calling run() command, e.g.:
>>> stocks = model.run(data_files="input_data.tab")
Several files can be passed by using a list. If the data information is not found in the first file, the next one will be used until finding the data values:
>>> stocks = model.run(data_files=["input_data.tab", "input_data2.tab", ..., "input_datan.tab"])
If a variables are defined in different files, to choose the specific file a dictionary can be used:
>>> stocks = model.run(data_files={"input_data.tab": ["data_var1", "data_var3"], "input_data2.tab": ["data_var2"]})
Note
Only tab and csv files are supported. They should be given as a table, with each variable in a column (or row) and the time in the first column (or first row). The column (or row) names can be given using the name of the variable in the original model or using python names.
Note
- Subscripted variables must be given in the Vensim format, one column (or row) per subscript combination. Example of column names for 2x2 variable:
subs var[A, C] subs var[B, C] subs var[A, D] subs var[B, D]
Outputting various run information¶
The run() command has a few options that make it more useful. In many situations we want to access components of the model other than merely the stocks - we can specify which components of the model should be included in the returned dataframe by including them in a list that we pass to the run() command, using the return_columns keyword argument:
>>> model.run(return_columns=['Teacup Temperature', 'Room Temperature'])
Teacup Temperature Room Temperature
0.000 180.000000 70
0.125 178.625000 70
0.250 177.267188 70
0.375 175.926348 70
0.500 174.602268 70
... ... ...
29.500 75.651312 70
29.625 75.580671 70
29.750 75.510912 70
29.875 75.442026 70
30.000 75.374001 70
[241 rows x 2 columns]
If the measured data that we are comparing with our model comes in at irregular timestamps, we may want to sample the model at timestamps to match. The run() function provides this functionality with the return_timestamps keyword argument:
>>> model.run(return_timestamps=[0, 1, 3, 7, 9.5, 13, 21, 25, 30])
Characteristic Time Heat Loss to Room Room Temperature Teacup Temperature FINAL TIME INITIAL TIME SAVEPER TIME STEP
0.0 10 11.000000 70 180.000000 30 0 0.125 0.125
1.0 10 9.946940 70 169.469405 30 0 0.125 0.125
3.0 10 8.133607 70 151.336071 30 0 0.125 0.125
7.0 10 5.438392 70 124.383922 30 0 0.125 0.125
9.5 10 4.228756 70 112.287559 30 0 0.125 0.125
13.0 10 2.973388 70 99.733876 30 0 0.125 0.125
21.0 10 1.329310 70 83.293098 30 0 0.125 0.125
25.0 10 0.888819 70 78.888194 30 0 0.125 0.125
30.0 10 0.537400 70 75.374001 30 0 0.125 0.125
Retrieving a flat DataFrame¶
The subscripted variables, in general, will be returned as xarray.DataArray in the output pandas.DataFrame. To get a flat dataframe, set flatten_output=True when calling the run() method:
>>> model.run(flatten_output=True)
Storing simulation results on a file¶
Simulation results can be stored as .csv, .tab or .nc (netCDF4) files by defining the desired output file path in the output_file argument, when calling the run() method:
>>> model.run(output_file="results.tab")
If the output_file is not set, the run() method will return a pandas.DataFrame.
For most cases, the .tab file format is the safest choice. It is preferable over the .csv format when the model includes subscripted variables. The .nc format is recommended for large models, and when the user wants to keep metadata such as variable units and description.
Note
PySD includes helper functions to export .nc file contents to .csv or .tab files. See Exporting netCDF data_vars to csv or tab for further details.
Warning
.nc files require netCDF4 library which is an optional requirement and thus not installed automatically with the package. We recommend using netCDF4 1.6.0 or above, however, it will also work with netCDF4 1.5.0 or above.
Setting parameter values¶
In some situations we may want to modify the parameters of the model to investigate its behavior under different assumptions. There are several ways to do this in PySD, but the run() method gives us a convenient method in the params keyword argument.
This argument expects a dictionary whose keys correspond to the components of the model. The associated values can either be constants, or pandas.Series whose indices are timestamps and whose values are the values that the model component should take on at the corresponding time. For instance, in our model we may set the room temperature to a constant value:
>>> model.run(params={'Room Temperature': 20})
Alternately, if we want the room temperature to vary over the course of the simulation, we can give the run() method a set of time-series values in the form of a pandas.Series, and PySD will linearly interpolate between the given values in the course of its integration:
>>> import pandas as pd
>>> temp = pd.Series(index=range(30), data=range(20, 80, 2))
>>> model.run(params={'Room Temperature': temp})
If the parameter value to change is a subscripted variable (vector, matrix…), there are three different options to set the new value. Supposse we have ‘Subscripted var’ with dims ['dim1', 'dim2'] and coordinates {'dim1': [1, 2], 'dim2': [1, 2]}. A constant value can be used and all the values will be replaced:
>>> model.run(params={'Subscripted var': 0})
A partial xarray.DataArray can be used. For example a new variable with ‘dim2’ but not ‘dim1’. In that case, the result will be repeated in the remaining dimensions:
>>> import xarray as xr
>>> new_value = xr.DataArray([1, 5], {'dim2': [1, 2]}, ['dim2'])
>>> model.run(params={'Subscripted var': new_value})
Same dimensions xarray.DataArray can be used (recommended):
>>> import xarray as xr
>>> new_value = xr.DataArray([[1, 5], [3, 4]], {'dim1': [1, 2], 'dim2': [1, 2]}, ['dim1', 'dim2'])
>>> model.run(params={'Subscripted var': new_value})
In the same way, a pandas.Series can be used with constant values, partially defined xarray.DataArray or same dimensions xarray.DataArray.
Note
Once parameters are set by the run() command, they are permanently changed within the model. We can also change model parameters without running the model, using PySD’s set_components() method, which takes the same params dictionary as the run() method. We might choose to do this in situations where we will be running the model many times, and only want to set the parameters once.
Note
If you need to know the dimensions of a variable, you can check them by using get_coords() method:
>>> model.get_coords('Room Temperature')
None
>>> model.get_coords('Subscripted var')
({'dim1': [1, 2], 'dim2': [1, 2]}, ['dim1', 'dim2'])
this will return the coords dictionary and the dimensions list, if the variable is subscripted, or ‘None’ if the variable is an scalar.
Note
If you change the value of a lookup function by a constant, the constant value will be used always. If a pandas.Series is given the index and values will be used for interpolation when the function is called in the model, keeping the arguments that are included in the model file.
If you change the value of any other variable type by a constant, the constant value will be used always. If a pandas.Series is given the index and values will be used for interpolation when the function is called in the model, using the time as argument.
If you need to know if a variable takes arguments, i.e., if it is a lookup variable, you can check it by using the get_args() method:
>>> model.get_args('Room Temperature')
[]
>>> model.get_args('Growth lookup')
['x']
Setting simulation initial conditions¶
Initial conditions for our model can be set in several ways. So far, we have used the default value for the initial_condition keyword argument, which is ‘original’. This value runs the model from the initial conditions that were specified originally in the model file. We can alternately specify a tuple containing the start time and a dictionary of values for the system’s stocks. Here we start the model with the tea at just above freezing temperature:
>>> model.run(initial_condition=(0, {'Teacup Temperature': 33}))
The new value can be a xarray.DataArray, as explained in the previous section.
Additionally, we can run the model forward from its current position, by passing initial_condition=‘current’. After having run the model from time zero to thirty, we can ask the model to continue running forward for another chunk of time:
>>> model.run(initial_condition='current',
return_timestamps=range(31, 45))
The integration picks up at the last value returned in the previous run condition, and returns values at the requested timestamps.
There are times when we may choose to overwrite a stock with a constant value (ie, for testing). To do this, we just use the params value, as before. Be careful not to use ‘params’ when you really mean to be setting the initial condition!
Querying current values¶
We can easily access the current value of a model component using square brackets. For instance, to find the temperature of the teacup, we simply call:
>>> model['Teacup Temperature']
If you try to get the current values of a lookup variable, the previous method will fail, as lookup variables take arguments. However, it is possible to get the full series of a lookup or data object with get_series_data() method:
>>> model.get_series_data('Growth lookup')
Copying a model¶
Sometimes, you may want to run several versions of a model. For this purpose, copying an already-loaded model to make changes while keeping an untouched one is useful. The copy() method will help do that; it will load a new model from the translated file and apply to it the same changes that have been applied to the original model (modifying components, selecting submodels, etc.). You can also load a copy of the source model (without applying) any change setting the argument reload=True.
Warning
The copy function will load a new model from the file and apply the same changes to it. If any of these changes have replaced a variable with a function that references other variables in the model, the copy will not work properly since the function will still reference the variables in the original model, in which case the function should be redefined.